RETROVIRUS REPLICATION
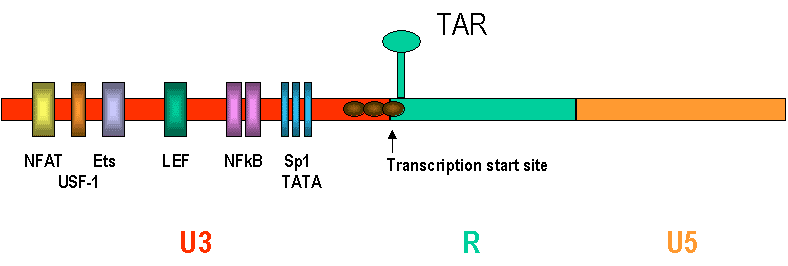
The long terminal repeat in the DNA proviral form of the retrovirus consists of three parts U3, R and U5. Originally, U3 was at the 3' end of the RNA form of the virus while U5 was at the 5' end. During formation of the DNA form, U3 and U5 become duplicated and transposed. This results in the site of initiation of RNA synthesis by RNA polymerase II being at the U3-R interface and the termination / poly A addition site being at the R-U5 interface. As a result copying of the DNA into RNA yields an RNA genome faithful (almost) to that of the infecting virus.
The LTR contains a number of important cis-acting elements that are called promotors that stimulate transcription. These are transcription factor binding sites. In the HIV U3 promotor region there is:
In the R region of the LTR there is also a region called TAR (transactivation response element)

Fig 1. The HIV LTR Image adapted from Freed EO. Somat. Cell Nol. Genet. 26, 13, 2001
The TAR is located in the first 57 nucleotides of the R region and is a stem-loop structure that binds to the protein TAT. Since it is downstream from the beginning of the R region (where the transcription start site is located), it is also present in the nascent RNA. This structure is important for transcription. TAT protein, which is translated from RNAs derived by splicing of the primary transcript and which is made at a basal rate after infection, is made in the cytoplasm and imported into the nucleus. Here it binds to the TAR region of the nascent RNA and stimulates further transcription. The TAT protein can also bind to the DNA provirus. In the absence of TAT, small RNAs are made that start at the U3/R interface and terminate in the TAR region. In the presence of TAT, the suppression is suppressed leading to transcription through the hairpin TAR region to the second R/U5 interface.
In the U3 region, there is an NFkB (nuclear factor kappa-b) transcription factor binding site (figure 1) which is important in regulation of HIV-1 during activation of T-cells. When a T cell that harbors an HIV-1 provirus is activated (by a mitogen or antigen), the phosphorylation and subsequent rapid degradation of an NFkB inhibitory cytoplasmic protein called I kappa B occurs. I kappa B kept the NFkB protein in the cytoplasm but after I kappa B degradation the NFkB moves to the nucleus where it binds to the LTR and activates transcription.
Like TAT, NFkB functions in both initiation of transcription and elongation. In initiation, NFkB alters the structure of the chromatin. DNA is bound by histones and other proteins into nucleosomes and this occurs in the integrated viral DNA as well as in the normal cellular chromatin. There are two nucleosomes in the HIV-1 LTR region (Nuc-0 and Nuc-1). Nuc-1 is at the transcription initiation site, that is at the U3-R interface. The proteins of the nucleosome inhibit transcription by limiting accessibility of the start site. Note that the NFkB and Sp1 sites are not in the regions of the LTR that are parts of the nucleosomes (figure 2). NFkB recuits other proteins (among which are CRB and p300 and p160, co-activator proteins). These have histone-specific acetyl transferase activity and hyperaceylate hsitones thereby disrupting the nucleosome and activating transcription at that site.
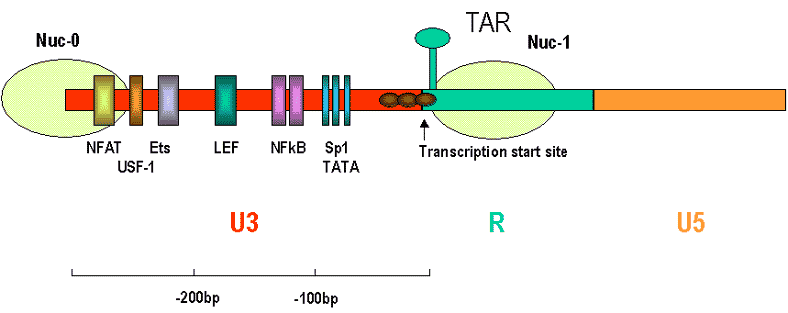
Fig 2. Nucleosomes in the HIV LTR Image adapted from Marzio and Giacca, 1999)
Not only can NFkB activate transcription but it can also regulate elongation of the mRNA in HIV-1 since it is observed that when NFkB is activated the proportion of the HIV-1 RNA transcripts that are full length is increased. Thus, a proportion of the RNA polymerases appear to function better in elongation. The p65 subunit of NfkB is responsible for this effect.
TAT protein regulates HIV-1 transcription by enhancing elongation of the transcripts after binding to the TAR region. It is essential that the TAR region is downstream of the HIV promotor region and must be in the correct orientation.