Dr. Margaret Hunt
MEDICAL MICROBIOLOGY, MBIM 650/720
Reading: Murray et al., Microbiology, 3rd Ed.: Ch. 6 and appropriate parts of Ch. 54 (Picornaviruses), Ch. 58 (Rhabdoviruses), Ch. 55 (Paramyxoviruses), Ch. 56 (Orthomxyoviruses), Ch. 57 (Reoviruses), Ch. 48 (Vial diagnosis)

VIROLOGY - LECTURE 4
RNA VIRUS REPLICATION STRATEGIES
TEACHING OBJECTIVES
Descriptive analysis of the replicative strategies employed by animal RNA viruses
Identification of virus prototypes associated with different RNA virus replication schemes
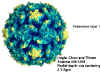
 Structure of Polio Type 1 Mahoney. X-ray data from Hogle et al.(Harvard Univ.), PDB entry 2PLV, rendered with GRASP (A.Nicholls, Columbia Univ.). Courtesy of
Dr Sgro and the Institute for Molecular
Virology, Univ. of Wisconsin (used with permission)
Structure of Polio Type 1 Mahoney. X-ray data from Hogle et al.(Harvard Univ.), PDB entry 2PLV, rendered with GRASP (A.Nicholls, Columbia Univ.). Courtesy of
Dr Sgro and the Institute for Molecular
Virology, Univ. of Wisconsin (used with permission)
RNA VIRUS REPLICATION - GENERAL
STRATEGIES
a) RNA VIRUSES WHICH DO NOT COPY THEIR RNA INTO DNA:
These viruses need an RNA-dependent RNA-polymerase to replicate their RNA, but animal cells do not seem to possess such an enzyme. Therefore, this type of animal RNA virus needs to code for an RNA-dependent RNA polymerase.
No viral proteins can be made until
viral messenger RNA is available. Thus, the nature of the RNA in the
virion affects the strategy of the virus.
i. plus-stranded RNA viruses
In these viruses, the virion (genomic) RNA is the same sense as mRNA and so functions as mRNA. This mRNA can be translated immediately upon infection of the host cell
Examples:
poliovirus (picornavirus)
togaviruses
flaviviruses
ii. negative-stranded RNA viruses
The virion RNA is negative sense (complementary to mRNA) and must therefore be copied into the complementary plus-sense mRNA before proteins can be made. Thus, besides needing to code for an RNA-dependent RNA-polymerase, these viruses also need to package it in the virion so that they can make mRNAs upon infecting the cell.
Examples:
influenza virus (orthomyxovirus)
measles virus, mumps virus, (paramyxoviruses)
rabies virus (rhabdovirus)iii. double-stranded RNA viruses
The virion (genomic) RNA is double stranded and so cannot function as mRNA; thus these viruses these also need to package an RNA polymerase to make their mRNA after infection of the host cell.
Example:
rotaviruses (belong to reovirus family)
b) RNA VIRUSES WHICH COPY THEIR RNA INTO DNA:
These are the retroviruses. In this case, their virion RNA, although plus-sense, is not released in the cytoplasm, and so does not function as mRNA immediately upon infection. Instead it serves as a template for reverse transcriptase and is immediately copied into DNA. Reverse transcriptase is not available in the cell, and so these viruses need to code for this enzyme and package it in virions.
THE TRANSLATION PROBLEM
Eucaryotic host cell translation protein synthesis machinery in general uses monocistronic mRNAs and so there is a problem in making more than one type of protein from a single mRNA.
RNA viruses have several solutions to this problem:
i. The virus makes multiple monocistronic mRNAs
ii. The virus makes primary transcripts which are processed by the host splicing machinery to give more than one monocistronic RNA
iii. The viral mRNA acts as a monocistronic transcript, a large polypeptide (called a polyprotein) is made which is then cleaved into separate proteins - Thus, one initial translation product is processed to give rise to multiple proteins. This happens, for example, in picornaviruses
GENOME SIZE OF RNA VIRUSES
RNA viruses tend to have a relatively small genome (although virion size may not necessarily be small). This is probably because the lack of RNA error correction mechanisms puts a limit on the size of RNA genomes.
The result of having a small genome is that RNA viruses tend to code for only a few proteins. These will include a polymerase which can copy RNA into a complementary nucleic acid (either RNA or, as in the case of retroviruses, DNA) and a viral attachment protein.
 Polio virus © 1999 Dr
J-Y
Sgro, University of Wisconsin Biotechnology Center, Molecular Graphics Resources.
Used with permission
Polio virus © 1999 Dr
J-Y
Sgro, University of Wisconsin Biotechnology Center, Molecular Graphics Resources.
Used with permission
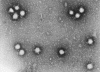
 Polio virus x 350,000 © Dr
Dennis Kunkel, University of Hawaii. Used with permission
Polio virus x 350,000 © Dr
Dennis Kunkel, University of Hawaii. Used with permission
POSITIVE STRAND RNA VIRUSES
Examples:
picornaviruses
togaviruses
flaviviruses
1. PICORNAVIRUSES
PROPERTIES
Small (28nm), naked icosahedral viruses
(pico=very small)
RNA is single-stranded, plus sense, polyadenylated - functions as mRNA immediately
upon infection
Prototype member: poliovirus
ADSORPTION AND PENETRATION
A viral protein recognizes a receptor on
the host cell
membrane (important in tropism of virus).
It seems that binding to receptor alters capsid structure in some way.
RNA is released into cytoplasm - Details of this process remain unknown.
mRNA is now available for translation.
SYNTHESIS OF VIRAL PROTEINS
Poliovirus virion RNA functions as an mRNA but does not have the methylated cap structure typical of eucaryotic mRNAs - it has a "ribosome landing pad" (internal ribosome entry site, IRES) which enables ribosomes to bind without having to recognize a 5' methylated cap structure.
The mRNA is translated into a single polypeptide (polyprotein), which is cleaved.
Cleavage occurs before translation is complete, i.e. on nascent chain.
Cleavages are carried out by virally coded proteases.
 Adapted from Schaechter et al., Mechanisms of
Microbial Disease, 2nd Ed.
Adapted from Schaechter et al., Mechanisms of
Microbial Disease, 2nd Ed.Products of cleavage include:
An RNA polymerase (replicase)
Structural components of the virion
Proteases
RNA REPLICATION
We now have newly made viral proteins to support replication.
1. Viral RNA polymerase copies plus-sense genomic RNA into complementary minus-sense RNA:
This process needsVPg (or precursor containing VPg)
RNA polymerase (replicase)
Host proteinsVPg may act as a primer for RNA synthesis, this would explain why it is at the 5' end of all newly synthesized RNA molecules
2. New minus sense strands serve as template for new plus sense strands. Again polio RNA replicase and VPg are needed. VPg is linked to the 5' ends of the new plus sense strands (again, it probably functions as a primer).
New plus strand has three alternative fates:
i. It may serve as a template for more minus strands
ii. It may be packaged into progeny virions
iii. It may be translated into polyprotein (In this case VPg is removed prior to translation)
ASSEMBLY
When sufficient plus-sense progeny
RNA and virion proteins have accumulated, assembly begins.
Particles assemble with VPg-RNA
inside and 3 proteins in the capsid [VP0,1 and 3].
VP0 is then cleaved to VP2 and VP4
and these mature virions are infectious.
Virions are released following cell
lysis
NOTE: THE ENTIRE LIFE CYCLE OCCURS IN THE CYTOPLASM
 Rhabdovirus on a Fish Epithelial Cell
©
Dr
Dennis Kunkel
(used with permission)
Rhabdovirus on a Fish Epithelial Cell
©
Dr
Dennis Kunkel
(used with permission)NON-SEGMENTED NEGATIVE STRAND VIRUSES
Examples:
Rhabdoviruses
Paramyxoviruses
 Structure of a typical rhabdovirus
Structure of a typical rhabdovirus1. RHABDOVIRUSES
Example: Rabies virus. The most intensively studied member is vesicular stomatitis virus.
RNA:
single stranded
negative (minus) sense
codes for 5 proteins
NOTE: THE ENTIRE LIFE CYCLE OCCURS IN THE CYTOPLASM
ATTACHMENT, PENETRATION AND
UNCOATING
The virus adsorbs to cell surface.
G (Glycoprotein) is attachment protein which binds to receptor on host cell.
The attached virus is endocytosed via clathrin coated pits.
The membrane of virus fuses with the endosome
membrane (acid pH of endosome is important because an acid pH is required for fusion to
occur).
The nucleocapsid is released into
cytoplasm.
TRANSCRIPTION:
'Transcription' is used in this
context to refer to synthesis of mRNAs.
Complete uncoating of the nucleocapsid
is not necessary - the virion RNA polymerase can copy virion RNA when it is in
the nucleocapsid form. This is an advantage in that genomic RNA is therefore
somewhat protected from ribonucleases.
There is one monocistronic mRNA for
each of the five virally coded proteins.
The mRNAs are capped, methylated,
and polyadenylated.
Since this is a cytoplasmic,
negative-sense RNA virus, the enzymes for mRNA synthesis and modification are
packaged in the virion.
TRANSLATION
mRNAs are translated on host
ribosomes.
All 5 proteins made.
There is no distinction between early and late functions.
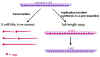
RNA REPLICATION
RNA replication is the process by
which new copies of genome-length RNAs are made.
RNA replication occurs in the
cytoplasm and is carried out by the viral RNA polymerase.
The full length plus strand is
coated with nucleocapsid protein as it is made (mRNAs are not coated with this
protein, which would interfere with the host protein translation machinery).
The new positive strand is copied into full
length minus strand, which is also coated with nucleocapsid protein as it is
made.
(Note: since the viral RNA
polymerase synthesises mRNAs (transcription) and full-length RNA
(replication), it is also sometimes called a transcriptase or a replicase , such
names just focus on the different aspects of the polymerase activity.)
New negative strands may:
i. be used as templates for the synthesis of more full length plus strands
ii. be used as templates for the synthesis of more mRNAs
iii. be packaged into virions

ASSEMBLY
The virus consists of two "modules" - the envelope and the nucleocapsid:
Envelope
Transmembrane proteins are made on
ribosomes bound to the endoplasmic reticulum. They are
inserted into the endoplasmic reticulum membrane as they are made. The
transmembrane proteins are glycosylated in the endoplasmic reticulum and pass
through the Golgi body where substantial modification of the carbohydrate chains
occurs. They are then transported, in vesicles, to the appropriate
cell membrane; in the case of vesicular stomatitis virus, this is the plasma
membrane.

Nucleocapsid
Synthesis of the nucleocapsid was described above. Once nucleocapsids are formed, the RNA polymerase associates with them - and the complex is now ready to be packaged.
Nucleocapsids bud out through modified areas of membrane which contain G and M proteins. The M (matrix) protein is involved in assembly - it interacts with patches of G in the membrane and with nucleocapsids.
 Paramyxovirus © Dr
Linda
Stannard, University of Cape Town, South Africa
(used with permission)
Paramyxovirus © Dr
Linda
Stannard, University of Cape Town, South Africa
(used with permission)
Paramyxoviruses:
Pleomorphic
Negative-sense, nonsegmented RNA, helical nucleocapsid
Enveloped
Envelope contains two virally coded glycoproteins: The F protein and the attachment proteinThe F protein has fusion activity.
The attachment protein binds to receptors on the host cell, this protein may have:hemagglutinating activity and neuraminidase activity (HN protein)
or hemagglutinating activity alone (H protein)
or neither (G protein).
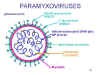 Structure of a typical paramyxovirus
Structure of a typical paramyxovirus|
PARAMYXOVIRUS FAMILY |
||
|
GENUS |
GLYCOPROTEIN |
TYPICAL MEMBERS |
|
Paramyxovirus genus |
HN |
mumps, parainfluenza viruses |
|
Morbillivirus genus |
H |
measles |
|
Pneumovirus genus |
G |
respiratory syncytial virus |
Hemagglutination is easy to test for in the clinical laboratory and is used in diagnosis
involves the agglutination of red blood cells. It relies on the ability of a virus to bind to receptors on red blood cells. Since viruses have multiple attachment proteins per virion, they can bind to more than one red blood cell and so they can serve to link red blood cells into a network. Inactivated virus can still hemagglutinate as long as its attachment proteins are intact.Hemagglutination
If someone has antibodies to a viral hemagglutinin, then the serum of that person will inhibit agglutination by the virus to which they have antibodies - but not by other hemagglutinating viruses. This can be used to determine which hemagglutinating virus a person has been exposed to.
Hemadsorption relies similarly on the ability to bind red blood cells. If a virus puts a viral attachment protein into the plasma membrane of the infected cell which can bind to receptors on red blood cells, then the infected cell will bind red blood cells. This may enable virally infected cells to be detected at an early stage in infection, and may enable the detection of viruses which do not visibly damage the cell.

ADSORPTION AND PENETRATION
The H(N)/G protein recognizes receptors on cell surface.
The F protein facilitates fusion between membranes at physiological pH, so although paramyxoviruses can be taken up by coated pits, they also often enter the cell by direct fusion with the plasma membrane. Hence syncytia can be formed in paramyxovirus infections.
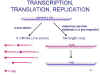
TRANSCRIPTION, TRANSLATION, REPLICATION OF RNA
Events inside the cell are very
similar to rhabdoviruses:
Viral multiplication occurs in the cytoplasm.
RNA polymerase and RNA modification enzymes are packaged in the virion.
The RNA polymerase does not need a fully uncoated nucleocapsid.
Viral mRNAs are transcribed; these are capped, methylated and polyadenylated
The viral mRNAs are translated to give viral proteins.
There is no distinction between early and late functions in gene expression.
Viral RNA replication involves full length plus strand synthesis. This is used as a template for full length minus strand. Both full length strands are coated with nucleocapsid protein as they are made.
New full length minus strands may serve as templates for replication, or templates for transcription, or they may be packaged into new virions.

ASSEMBLY
Both viral glycoproteins [i.e.
attachment protein and F (fusion) protein] are translated as transmembrane
proteins and transported to the cell plasma membrane.
M (matrix) protein enables nucleocapsids to interact with the regions of the
plasma membrane which have the glycoproteins inserted.
Virus buds out through membrane.
ROLE OF THE NEURAMINIDASE
In those paramyxoviruses which have it, the neuraminidase may facilitate release. In these viruses, sialic acid appears to be an important part of the receptor. The neuraminidase removes sialic acid (neuraminic acid) from the cell surface. Thus, since sialic acid will have been largely removed from the cell surface and the progeny virions, neither will have functional receptors, so progeny virions will not stick to each other or to the cell they have just budded out from (or any other infected cell). They will therefore be able to diffuse away until they meet an uninfected cell.
The neuraminidase may also help during infection since, if the virus binds to sialic acid residues in mucus, it would not be able to bind to a receptor on a cell and infect that cell. But if the sialic acid in the mucus is eventually destroyed, the virus will be freed and may then reach a receptor on the cell surface.
ACTIVATION OF THE F PROTEIN
The F protein needs to be cleaved before it can function in facilitating fusion when the virus binds to another cell. This is a late event in maturation.
 Orthomyxovirus (Influenza A) © Dr Linda Stannard,
University of Cape Town, South Africa
Orthomyxovirus (Influenza A) © Dr Linda Stannard,
University of Cape Town, South Africa
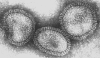 Orthomyxovirus (Influenza A) © Dr Linda Stannard,
University of Cape Town, South Africa
Orthomyxovirus (Influenza A) © Dr Linda Stannard,
University of Cape Town, South Africa
SEGMENTED NEGATIVE STRAND VIRUSES
Examples:
Orthomyxoviruses
Bunyaviruses (include Hantavirus genus)
Arenaviruses
ORTHOMYXOVIRUSES
There are three groups of influenza virus: A, B and C. Influenza A virus is most intensively studied.
Pleomorphic virions
Negative-sense, single-stranded RNA
RNA genome is SEGMENTED, eight
segments in influenza A
Helical nucleocapsid
Virions contain RNA polymerase
Enveloped, two membrane glycoproteins:
HA - hemagglutinin - attachment and
fusion protein
NA - neuraminidase - important in
release
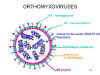 Structure of a typical orthomyxovirus
Structure of a typical orthomyxovirusVirus adsorbs to receptors on the cell surface.
Virus is engulfed by coated pits.
At acid pH of an endosome, HA undergoes a conformational change and fusion occurs.
Nucleocapsids are released to cytoplasm.

TRANSCRIPTION, TRANSLATION AND REPLICATION
Nucleocapsids are transported into the nucleus. mRNA synthesis and replication of viral RNA occurs in the nucleus. This is very unusual for an RNA virus.
Influenza virus has an unusual mechanism for acquiring a methylated, capped 5'end to its mRNAs:
A viral endonuclease (packaged in the influenza virus) snips off the 5'end of a host capped, methylated mRNA about 13-15 bases from the 5' end and uses this as a primer for viral mRNA synthesis - hence all flu mRNAs have a short stretch at the 5' end which is derived from host mRNA.
The viral RNA polymerase (transcriptase)
copies the template into complementary plus sense mRNA and adds a poly(A) tail.
Products of transcription:
The virus makes 8 primary transcripts: one per segment.
Two of these segments give rise to primary transcripts which can be alternatively spliced (since influenza virus RNA synthesis occurs in the nucleus, it has access to splicing machinery), each giving rise to two mature mRNAs. Thus a single segment can code for more than one protein since the virus has access to splicing machinery.
mRNAs are translated in the cytoplasm. Transmembrane proteins are moved to the plasma membrane while proteins needed for RNA replication go to the nucleus.
REPLICATION OF RNA
RNA replication occurs in the nucleus using a virus coded enzyme (this may be same as the RNA polymerase involved in
transcription of mRNAs, or a modified version).
A full length, exact
complementary copy of virion RNA is made - this plus sense RNA is probably coated with
nucleocapsid protein as it is made.
Full length plus strand RNA is then
used as a template for full-length minus strand synthesis; again the new minus
strand is probably coated with nucleocapsid protein as it is made.
New minus strands can be used as
templates for replication, mRNA synthesis, or packaged.
ASSEMBLY
This occurs at the plasma membrane.
Nucleocapsids are transported out
of the nucleus.
Envelope proteins are transported
via the Golgi body to the plasma membrane.
The M protein is involved - it interacts with both nucleocapsid and modified region of plasma membrane which
contains the glycoproteins HA and NA.
Virus buds out.
HA needs to be cleaved before it can promote fusion on the next round of infection. The requirement for cleavage affects which tissues can produce infectious virus.NOTE:
NA probably helps the virus leave the cell by removing sialic acid from receptors. NA may also help the virus penetrate mucus to reach epithelial cells of respiratory tract by enabling it to dissociate from sialic acid-containing receptors in the mucus by destroying them.
(The virus can infect cells because endocytosis presumably occurs before the receptors can be destroyed.)
There are similarities and differences between the Paramyxovirus family and the Orthomyxovirus family, members of both are enveloped, both contain negative sense, single stranded RNA, have helical nucleocapsids. However, the two families are very different. There is NO immunological relationship between the two families.
|
PROPERTY |
PARAMYXOVIRIDAE |
ORTHOMYXOVIRIDAE |
|
Genome |
non-segmented |
segmented |
|
RNA synthesis |
cytoplasmic |
nuclear |
|
Need for mRNA primer |
no |
yes |
|
Hemagglutinin,neuraminidase |
if both, part of same protein (HN) |
on 2 different proteins (HA,NA) |
|
Syncytia formation |
yes (F functions at at normal physiol. pH) |
no (HA functions at acid pH) |
 Mammalian Reovirus Virion Copyright
1999 Dr J-Y Sgro - University of Wisconsin Biotechnology Center, Molecular Graphics Resources
Mammalian Reovirus Virion Copyright
1999 Dr J-Y Sgro - University of Wisconsin Biotechnology Center, Molecular Graphics ResourcesDOUBLE STRANDED RNA VIRUSES
REOVIRUS FAMILY
Reovirus family include:
the members of the reovirus genus
the members of the rotavirus genus
the members of the orbivirus genus
Colorado tick fever virus
 Structure of a typical reovirus Adapted from Joklik et al. Zinsser
Microbiology 20th Ed.
Structure of a typical reovirus Adapted from Joklik et al. Zinsser
Microbiology 20th Ed.Icosahedral symmetry, multiple
shelled capsid
RNA is double stranded. There are 10-12 segments (depending on the genus of the Reovirus family
to which the virus belongs)
There are some significant
differences in the life cycle of members of the reovirus family and of the
rotavirus family. Due to their clinical importance in humans, we shall focus on
rotaviruses.
 Rotavirus (A double-capsid particle (left), and a single, inner, capsid (right))
Copyright Dr
Linda
Stannard, University of Cape Town, South Africa
Rotavirus (A double-capsid particle (left), and a single, inner, capsid (right))
Copyright Dr
Linda
Stannard, University of Cape Town, South AfricaROTAVIRUSES (rota = wheel)
ADSORPTION PENETRATION AND
UNCOATING
It is still not clear what exactly what
happens in vivo.
There appears to be a need for a protease to
generate an "intermediate sub-viral particle" (ISVP) before the virus can enter
the cytoplasm.
In vivo, the ISVPs are probably
generated by protease digestion in the GI tract.
A viral attachment protein,
probably at the vertices, recognizes host cell receptors.
The activated (ISVP) enters the
cytoplasm directly or via endocytosis.
TRANSCRIPTION AND TRANSLATION
Double stranded RNA does not
function as an mRNA and so the initial step is to make mRNA (transcription).
The mRNAs are made by virion-packaged RNA polymerase. It is also capped and methylated by virion packaged
enzymes. It is then
extruded from the capsid.

The mRNAs are translated. The resulting viral proteins assemble to form an immature capsid. (It is not known how the virus ensures that each particle gets one copy of the 11 different mRNAs.)
The mRNAs are packaged in the capsid and are now copied within the capsid to form double stranded RNAs.
More mRNAs are now made by the newly formed immature capsids.
More proteins are made and the immature capsids bud into the lumen of the endoplasmic reticulum. In doing so, they acquire a membrane, a transient envelope which is lost as they mature. This is a very odd feature of the rotaviruses.
RELEASE
Probably occurs via cell lysis.
![]() Return to the Virology section of Microbiology and Immunology On-line
Return to the Virology section of Microbiology and Immunology On-line
![]() Return to the Department of Microbiology and
Immunology Site Guide
Return to the Department of Microbiology and
Immunology Site Guide
This page
copyright 2001, The Board of Trustees of the University of South Carolina
This page last changed on Wednesday, May 28, 2003
Page maintained by Richard Hunt
URL: http://www.med.sc.edu:85/mhuntRNA-HOt.htm
Please report any problems to rhunt@med.sc.edu