|
x |
x |
|
 |
 |
|
INFECTIOUS
DISEASE |
BACTERIOLOGY |
IMMUNOLOGY |
MYCOLOGY |
PARASITOLOGY |
VIROLOGY |
|
|
VIROLOGY CHAPTER EIGHTEEN
HEPATITIS VIRUSES
Dr Richard Hunt
Professor
Department of Pathology, Microbiology and Immunology
University of South Carolina School of Medicine
|
|
EN
ESPANOL -
SPANISH |
|
|
|
|
Let us know what you think
FEEDBACK |
|
SEARCH |
|
|

 |
|
Logo image © Jeffrey
Nelson, Rush University, Chicago, Illinois and
The MicrobeLibrary |
|
I am grateful to Peniel Dimberu (Yale
University) for corrections to this page |
 Figure 1A
Figure 1A
Hepatitis A virus
CDC |
Several diseases of the liver,
collectively known as hepatitis, are caused by viruses. The viruses
involved, five of which have been reasonably well characterized, come from a
wide range of virus families.
- Hepatitis A virus is a
picornavirus, a small
single strand RNA virus
- Hepatitis B virus belongs to the hepadnavirus
family of double stranded DNA viruses (see below)
- Hepatitis C virus is a flavivirus, a
single stand RNA virus
- Hepatitis D which is also known as Delta
agent is a circular RNA that is more similar to a plant
viroid than a complete virus
- Hepatitis E, also an RNA virus, is similar to a calicivirus
-
Hepatitis G virus is a flavivirus. Its involvement
in human disease, if any, is obscure
For a
summary of the hepatitis viruses, see Table1.
|
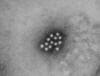 Figure 1B
Figure 1B
An electron micrograph of the Hepatitis A virus (HAV)
CDC - Betty Partin |
HEPATITIS A VIRUS
This picornavirus (figure 1) is the causative agent of
infectious hepatitis. Picornaviruses have a single strand,
3-polyadenylated, positive sense RNA genome surrounded by a naked (unenveloped)
icosahedral capsid that is around 28 nm in diameter (figure 2). At the 5 end of the
RNA strand is a viral protein called VPg. There is only one serotype of HAV.
|
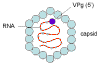 Figure 2
Figure 2
Hepatitis A virus - a picornavirus |
Replication
The virus binds to a receptor that is found on the surface of hepatocytes
and a few other cells. HAV cellular receptor 1 (havcr-1) has an ectodomain
that contains an N-terminal cysteine-rich immunoglobulin-like region,
followed by a mucin-like region that extends the immunoglobulin-like region well above
the cell surface. The immunoglobulin-like region is required for binding of HAV. The
virus spends its entire life in the cytoplasm where it replicates using a
virus-encoded RNA-dependent RNA polymerase. For further information on
picornavirus replication see Virology Section
Chapter Four.
|
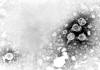 Figure 3A
Figure 3A
Transmission electron micrograph of hepatitis B virions, also known as Dane
particles
CDC/Dr. Erskine Palmer
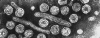 Figure 3B
Figure 3B
Hepatitis B virus © Dr Linda
Stannard, University of
Cape Town, South Africa. Used with permission
 Figure 3C
Figure 3C
Hepatitis B virus
CDC |
HEPATITIS B VIRUS
Human hepatitis B virus (figure 3) is the prototype virus of
the hepadnavirus family and causes serum hepatitis. HBV has a diameter of
about 40nm. It infects humans and chimpanzees but there are closely related
members of this family that infect other mammals and birds. HBV is a DNA virus and is enveloped. The
DNA is only partly double stranded and forms a circle of around 3,200 bases.
Although surrounded by a host cell-derived envelope, HBV is remarkably
stable to organic solvents. It is also heat- and pH-resistant. The genome is
associated with the P (polymerase) protein and this complex is, in turn,
surrounded by the core antigens (HBcAg and HBeAg). These two proteins have
most of their sequence in common and most of the HBeAg is secreted since it
is processed differently from the HBcAg and thus not assembled into progeny
virus. Embedded in the surface lipid bilayer is
the surface antigen (HBsAg). The HBsAg (Australia antigen) is made up of
three glycoproteins that are encoded by the same gene. The proteins are
translated in the same reading frame but start at a different AUG start
codon; thus, all have the same C-terminus. The largest protein is the L
protein (42kd) and contained within this is the M glycoprotein. The S
glycoprotein (27kD) is contained within the M protein. The HBsAg protein is
also secreted into the patients serum where it can be seen as spherical
(mostly self-associated S protein) or filamentous particles (also mostly S
protein but with some L and M). The former are smaller than the true virus
but the filaments can be quite large (several hundred nanometers). This
large amount of free HBsAg accounts for the inability to detect antibodies
against the protein early during infection (the so-called "window" between
the presence HBsAg (indicative of the presence of virus) and the presence of
anti-HBsAg).
The
glycoproteins on the virus surface contain antigenic determinants that are
group specific and type specific. Using these determinants, epidemiologists
identify eight subtypes of HBV. HBV virions are also known as Dane
particles.
|
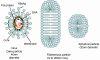 Figure 3D Hepatitis B virus. Dane particle and incomplete particles that are
found in patient's serum
Figure 3D Hepatitis B virus. Dane particle and incomplete particles that are
found in patient's serum
 Figure 3E
Figure 3E
Hepatitis B virus structure © Dr Linda
Stannard, University of
Cape Town, South Africa. Used with permission
 i
i
 ii
ii
 iii
iii
Figure 4A
Hepatitis B replication
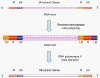 Figure 4B
Figure 4B
Genome replication in retroviruses
 i
i
 ii
ii
Figure 4C
Genome replication in hepadnaviruses
|
Replication
HBV has a very curious way of replicating itself since (figure 4A), although it is a DNA
virus, it uses a RNA proviral intermediate that has to be copied back to
DNA. The copying of RNA to DNA is not a normal function of an uninfected
cell but is found in retroviruses that also have an RNA genome and a DNA
intermediate that gets integrated into host cell chromosomes. For the
purpose of copying RNA to DNA, retroviruses and HBV have a virally-encoded
DNA polymerase (P) called reverse transcriptase.
After the HBV has attached to the cell surface
receptor (which has yet to be identified but may be a member of the
ovalbumin family of serine protease inhibitors), the viral membrane fuses
with the cell membrane releasing the core into the cytoplasm. The core
proteins dissociate from the partially double stranded DNA. DNA polymerase
now completes the DNA so that it is completely double stranded. This is done
by the virally-encoded polymerase in the cytoplasm that is one of the core
proteins (whereas the cells DNA polymerase is in the nucleus). The double
stranded DNA enters the nucleus and the ends are ligated by host enzymes so
that the virus is in the form of a circular episome. The viral DNA
associates with host nuclear histones and is transcribed by cellular RNA
polymerase II into mRNAs. In contrast to the situation with retroviruses, however,
the DNA form of HBV is usually not integrated into cellular DNA; rather it
is found as an independent episome. This is because, unlike retroviruses,
hepadnaviruses have no integrase activity. However,
integrated parts of the HBV genome are found in the chromosomes of many
hepatocellular carcinoma patients.
Four mRNAs are made from the HBV genome. The
host cell RNA polymerase interacts with four promoters but transcription always ceases
at the same polyadenylation site so that the overlapping mRNAs have a common
3 terminus. One of these mRNAs is slightly longer than the DNA sequence
because of the polyadenylation at one end and a repeated region. This is the full length c-RNA
that will be the template for the genome. The full length messenger RNA
codes for the polymerase and core HBcAg and HBeAg proteins. The latter are
very similar because they are translated in the same reading frame from two
different start codons. Two smaller mRNAs (2.4 and 2.1 bases) which overlap
code for the surface glycoproteins. There is also a small mRNA of 700 bases
that codes for a protein that is a protein kinase and is a transactivator of
transcription.
In the cytoplasm, the full-length (3,500 base)
positive strand c-RNA is encapsidated by core proteins. Inside the core, the
RNA is transcribed to minus strand DNA by the same DNA polymerase (reverse
transcriptase) that completed the double stranded DNA and,
at the same time, the RNA is degraded by a ribonuclease H that is also part
of the reverse transcriptase. Unlike the reverse transcriptase of the
retroviruses, the HBV reverse transcription reaction does not require a tRNA
primer. Rather, the polymerase itself acts as a primer and remains
covalently attached to the 5 end of the negative strand DNA. A host cell
chaperone protein, heat shock protein 90, is also necessary. The chaperone
associates with the reverse transcriptase allowing it to fold into an active
conformation.
The virus now buds through the endoplasmic
reticulum and/or Golgi Body membranes (or perhaps a novel pre-Golgi
compartment) of the host cell from which it acquires HBsAg. At this stage or
later, the minus stand of DNA is partly transcribed into a plus strand. When
the viral DNA polymerase is used to transcribe RNA to DNA, it is acting as a
reverse transcriptase similar to that found in retroviruses; in fact, HBV
DNA polymerase and retroviral reverse transcriptase are very similar, and
may have evolved from a common ancestor.
Virus particles that contain RNA or DNA at
various stages of replication can be found in the bloodstream suggesting that nucleic acid
replication is not tightly controlled with the passage out of the cell. In
addition, empty envelopes containing the envelope proteins embedded in a
lipid bilayer are continuously being shed.
RNA polymerase problem
There is a distinct problem posed by using host cell RNA polymerase II to
transcribe a DNA viral genome to an RNA form (See section on
retroviruses).
The normal function of RNA polymerase II is to transcribe a gene into
messenger RNA for subsequent translation into protein. In the mRNA, all that
is required is the information to make the protein. In the DNA gene,
additional information is present that is needed to make the RNA. This extra
information (that is not transcribed into RNA) includes the promoter (the
site at which the RNA polymerase binds), the enhancers that are up- and
down- stream of the region transcribed to mRNA and the polyadenylation site.
Thus, a messenger RNA is smaller than the DNA gene, even if there are no
introns.
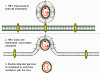
Retroviruses overcome the loss of promoter/enhancer information as a result
of using RNA polymerase II transcription by carrying internal copies of the
promoter and enhance regions (these are the U3 and U5 sequences
respectively). They duplicate their internal U3 promoter sequence and
transpose it to the opposite end when the DNA is transcribed from RNA.
Similarly, the enhancers and other 3 information are stored internally (as
U5) and transposed to the other end. These events give rise to the long
terminal repeats (LTRs) that
are only found in the DNA form of the virus. When the RNA polymerase
recognizes the promoter in the U3 region, it finds the transcription
initiation site at the border between the U3 and R and starts transcribing
at the beginning of the R region. This leads to a faithful copy of the
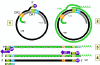
original RNA as the terminal U3 and U5 are lost (figure 4B).
The same problem occurs in hepadnaviruses which also have a DNA form of
their genome that is copied to RNA by host cell RNA polymerase II before
copying the RNA back to DNA using reverse transcriptase. However, the
mechanism is different; in this case, the DNA form of the virus is smaller
than the RNA form, quite the opposite of what occurs in the retroviruses.
The hepadnaviruses are small DNA viruses and, in contrast to the
retroviruses, it is the DNA that is packaged into the viral particle. This
DNA is copied to RNA in the infected cell by RNA polymerase II and the
resulting RNA is copied back to DNA by reverse transcriptase in the maturing
virus particle.
In the viral particle, the DNA is only partially double stranded. The
negative strand is complete, though not ligated into a circle. There are
free 5 (with an attached reverse transcriptase protein molecule) and 3
ends. The DNA is in the form of a relaxed circle because it is hybridized to
a partial copy of the positive strand. The DNA contains two direct repeats
(DR1 and DR2). DR1 is close to the 5 end of the negative strand and DR2 is
close to the 5 end of the partial positive strand.
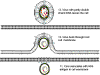
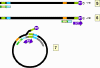
On entering the nucleus, the negative strand is ligated to form a covalently
closed circle. This is then copied by host RNA polymerase II. The polymerase
starts about 6 bases to the left (in figure 4Ci-2) of the DR1 and proceeds
(clockwise in figure 4Ci-2) around the circle past both the initiation site
and the DR1 and stops at the termination/poly A site (light blue) that is a
little further downstream. The RNA becomes polyadenylated. The RNA copy is
therefore larger than the covalently closed circular DNA (compare the
situation in retroviruses) because the DR1 region has been duplicated and
poly A has been added.
This RNA moves to the cytoplasm where encapsidation by viral proteins
occurs. There is an encapsidation signal at the 5 end of the RNA and thus
only one RNA molecule is found in each virion (compare the situation in
retroviruses). Now, in the virus particle itself, the RNA is copied to DNA
using reverse transcriptase. All DNA polymerases need a primer and in the
case of the retroviruses this is a host cell tRNA that is packed in the
virion. In the hepadnaviruses, the polymerase is packaged in the virion as
it is in the retroviruses, though there are fewer polymerase proteins per
virus particle in the hepadnaviruses. The reverse transcriptase is itself
the primer for the synthesis of the negative DNA strand and it remains
attached to the 5 end of the DNA via a tyrosine residue.
The DNA initiates on a hydroxyl group of the tyrosine using, as a template,
a region near the 5 end of the RNA (fig 4Ci-3). The polymerase copies
through the DR1 near the 5 end of the RNA and terminates at the end of the
RNA molecule. Next, a template exchange occurs in which the nascent negative
strand DNA moves to the DR1 near the 3 end (fig 4Ci-4). Why this is
necessary is obscure since the initiation could have occurred near the 3
DR1. From the 3 DR1, the DNA is extended accompanied by RNase H digestion
of the template RNA strand. Synthesis stops when the 5 end of the RNA is
reached (figure 4Ci-4). The negative strand is now terminally redundant. The
RNA is not completely destroyed and the last 15 or so nucleotides remain
(figure 4Cii-5) to serve as a primer for the second (positive) DNA strand
synthesis. This is translocated to the DR2 at the 5 end of the first DNA
stand (figure 4Cii-6). Extension continues to the 5 end of the first DNA
strand. There now occurs a switch of template in which the DR1 at the 5 end
of the negative strand is replaced by the DR1 at the 3 end so circularizing
the template (figure 4Cii-7). The reverse transcriptase now copies around
the circle for a variable distance to form the DNA that is found in mature
virus particles.
Carcinogenesis
It is clear that individuals who are HBsAg
positive are at a much higher risk of hepatocellular carcinoma than those
who are negative. In patients with chronic hepatitis, there is destruction
of hepatocytes as a result of the immune response to the virus. This results
in regeneration (by cell division) of liver cells that may ultimately cause
the cancer. Although the virus does not integrate during the course of
normal replication, parts of the HBV genome are found integrated into the
DNA of hepatocellular carcinoma patients. This may result in the activation
of a cellular proto-oncogene in much the same way as occurs in some
retrovirus-caused cancers; in fact, in most cases of woodchuck
hepatocellular carcinoma (a widely used model system), viral DNA is found
close to the myc or a similar proto-oncogene. Hepatocellular carcinoma takes
many years to develop and this may reflect the rarity of integration in the
absence of an integrase enzyme. The tumor that does develop is thus likely
to be clone of a single cell where this process has occurred. An HBV
protein called protein X is known to activate the src kinase and this may
also underlie HBV carcinogenesis. This protein may also interact with p53,
one of the cell's tumor suppressor genes.
|
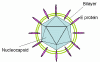 Figure 5
Figure 5
Hepatitis C structure
 Figure 6
Figure 6
Flavivirus polyprotein processing |
HEPATITIS C VIRUS
Hepatitis C is a flavivirus (of which yellow fever
is the prototype) that causes non-A, non-B hepatitis. Flaviviruses (figure
5) are
icosahedral, positive strand RNA viruses and gain an envelope from their
host cell. The virus particle is about 30 to 60nm across. The genome of
9,600 bases codes for ten proteins. In many ways, the flaviviruses are
similar to picornaviruses with the prominent exception that they are
enveloped. The viral RNA does not have a 5 cap or 3 poly A tract.
Translation of the viral RNA is mediated by the internal ribosome entry site
(IRES).
There is one protein product from one open reading frame.
The hepatitis C virus polyprotein is cleaved by both a virally-encoded
protease activity and a cellular protease. The nascent protein contains a signal
sequence that results in the translating ribosome attaching to the
cytoplasmic surface of the endoplasmic reticulum. The envelope protein (E)
thus crosses and embeds in the membrane and the signal sequence is removed
by a cellular signal protease. This results in the remainder of the protein,
the core protein, becoming cytoplasmic. It is cut by two viral proteases.
The C-terminal domain of NS2 is a cysteine protease and cleaves at the
NS2/NS3 junction. Another protease (NS3/4A serine protease) cleaves the
remaining junctions.
Thus, the core protein is cut into NS1, NS2, NS3
and NS4 proteins. NS2 and NS4 are then cut again (to give NS2a, NS2b, NS4a
and NS4b)
HCV binds to either the CD81 antigen or low density
lipoprotein (LDL) receptor on hepatocytes via its E2 glycoprotein. There is
also some evidence that it may bind to glycosaminoglycans.
|
 Figure 7
Figure 7
Hepatitis Delta agent CDC |
HEPATITIS DELTA AGENT
Hepatitis D (figure 7) is a highly defective virus since it
cannot produce infective virions without the help of a co-infecting helper
virus. This helper virus is hepatitis B virus that supplies the HBsAg
surface protein. In budding out of the cell, HDV acquires a membrane
containing HBsAg. HDV is similar to a plant viroid in that it has a small
circular RNA genome (1,700 bases) but unlike the plant viroids, the RNA
encodes a protein called the delta antigen. This complexes with the RNA. The
RNA is single stranded negative sense and is a covalently closed circle.
Because of a large amount of base pairing, the RNA takes on a rod-like
structure (figure 8).
|
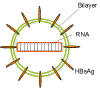 Figure 8
Figure 8
Hepatitis Delta agent - structure
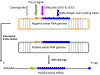 Figure 9
Figure 9
Hepatitis delta agent. Three RNA forms. Adapted from Wagner
and Hewitt.: Basic Virology. Blackwell Publishing |
HDV can only form an infectious
particle if the cell in which it replicates is co-infected with HBV
since the latter provides the surface HBsAg which is required for
reinfection of another cell. The HBsAg of HDV binds to the
same surface receptor as HBV and the virus fuses with the cell membrane. The
tropism of HDV is therefore the same as HBV. The
RNA genome is coated with delta antigen, the only
protein encoded by the RNA. The delta antigen, which is exposed when the
envelope is lost, has a nuclear localization
signal that targets the genome to the nucleus. Here the genome is copied by
host cell RNA polymerase II, the enzyme that normally makes mRNA. RNA
polymerase II is used by some other viruses to copy their genomes, for
example, the retroviruses, but in that case the polymerase copies DNA to
RNA (which is the normal function of the enzyme in the uninfected cell). In HDV replication, the polymerase is copying RNA to RNA. The negative
sense genomic RNA is copied to a positive strand that is also circular. The
genomic RNA can also be transcribed into a linear 5 capped and 3
polyadenylated mRNA which is smaller than the genomic RNA and contains the
small open reading frame from which the delta antigen is translated; or it
can be generated from the circular positive sense genomic-sized RNA by an
autocatalytic process that cleaves the RNA. Thus, the RNA is acting as a
ribozyme, that is a catalytic RNA (figure 9). Delta
antigen, translated from the mRNA has two forms that differ in size by 19
amino acids (195 compared to 214 residues). The formation of the large delta
antigen happens by a rather strange mechanism in which a host cell enzyme
called double stranded RNA-activated adenosine deaminase converts a UAG
(stop) codon into a UGG that allows translation to proceed to the next stop
codon. The small delta antigen is involved in the replication of the genome
but the larger form suppresses replication. This leads to the promotion of
viral particle assembly.
|
 Figure 10
Figure 10
Hepatitis E virus
CDC |
HEPATITIS E VIRUS
This virus (figure 10), which causes enteric non-A, non-B
hepatitis, seems to be related to the Caliciviruses but its classification
is undecided since the genome organization is not the same as that of the
Caliciviridae. In sequence, HEV is more similar to rubella which is a
Togavirus than to any Calicivirus. HEV is a small (approximately 34nm),
round, icosahedral, positive strand RNA virus that does not have an
envelope. It has a rather smooth surface but not as smooth a HAV. The genome
has a poly A tract and is capped at the 5 end. There are three open reading
frames that overlap; each is in a different coding frame. Based on sequence
motifs, open reading frame 1 (ORF1) appears to have several enzymic
activities. These may be involved in RNA capping, proteolysis and an
RNA-dependent RNA polymerase activity. ORF2 is the structural protein and
may be glycosylated. It appears to have a signal sequence suggesting that
its encoded protein may enter the endoplasmic reticulum. The third ORF codes
for a phosphoprotein of unknown function that interacts with the host cells
cytoskeleton. Not much is known about HEV replication but it is likely that
the positive strand RNA is copied to a negative strand intermediate by a
viral polymerase
|
| |
HEPATITIS G VIRUS
Hepatitis G virus is a flavivirus, like HCV to
which it is closely related. It is associated with some cases of acute or
chronic non-A, non-B, non-C, non-D, non-E hepatitis. Although it seems
common in human blood, it may not he a significant cause of hepatitis in
humans. |
| |
|
Table 1 |
| |
Hepatitis A |
Hepatitis B |
Hepatitis C |
Hepatitis Delta |
Hepatitis E |
| Virus
family |
Picornavirus |
Hepadnavirus |
Flavivirus |
Circular RNA
similar to plant viroid |
Similar to
Calicivirus |
| Nucleic
acid |
RNA (+
sense) |
DNA
(partially double strand) |
RNA (+
sense) |
RNA (-
sense) |
RNA (+
sense) |
| Disease
caused |
Infectious
hepatitis |
Serum
hepatitis |
Non-A, non-B
hepatitis |
|
Enteric
non-A, non-B hepatitis |
| Size |
~ 28nm |
~40nm |
30 - 60nm |
~ 40nm |
30 - 35 nm |
| Envelope |
No |
Yes |
Yes |
Yes |
No |
|
|
 Return to the Virology section of Microbiology and Immunology On-line
Return to the Virology section of Microbiology and Immunology On-line
This page last changed on
Friday, February 05, 2016
Page maintained by
Richard Hunt
|

 Figure 1A
Figure 1A Figure 1B
Figure 1B Figure 2
Figure 2 Figure 3A
Figure 3A Figure 3D Hepatitis B virus. Dane particle and incomplete particles that are
found in patient's serum
Figure 3D Hepatitis B virus. Dane particle and incomplete particles that are
found in patient's serum Figure 5
Figure 5 Figure 7
Figure 7 Figure 8
Figure 8 Figure 10
Figure 10









