| xx | x | ||||
 |
|||||
| BACTERIOLOGY | IMMUNOLOGY | MYCOLOGY | PARASITOLOGY | VIROLOGY | |
|
|
|
||||
| SPANISH | |||||
| TURKISH | |||||
| ALBANIAN | |||||
|
Let us know what you think FEEDBACK |
|||||
| SEARCH | |||||
|
|
|||||
|
|
|||||
|
TEACHING OBJECTIVES Introduction to animal virus genetics
|
GENERAL Viruses grow rapidly, there are usually a large number of progeny virions per cell. There is, therefore, more chance of mutations occurring over a short time period. The nature of the viral genome (RNA or DNA; segmented or non-segmented) plays an important role in the genetics of the virus. Viruses may change genetically due to mutation or recombination
MUTANTS
Some viral enzymes are not always essential and so we can isolate viable enzyme-deficient mutants; e.g. herpes simplex virus thymidine kinase is usually not required in tissue culture but it is important in infection of neuronal cells "Hot" mutants Attenuated mutants
|
||||
|
|
EXCHANGE OF GENETIC MATERIAL
|
||||
|
|
|
||||
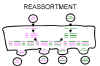 Figure 3 Reassortment of virus genome in segmented viruses
Figure 3 Reassortment of virus genome in segmented viruses |
If a virus has a segmented genome and if two variants of that virus infect a single cell, progeny virions can result with some segments from one parent, some from the other. This is an efficient process - but is limited to viruses with segmented genomes - so far the only human viruses characterized with segmented genomes are RNA viruses e.g. orthomyxoviruses, reoviruses, arenaviruses, bunya viruses. Reassortment may play an important role in nature in generating novel reassortants and has also been useful in laboratory experiments (figure 3). It has also been exploited in assigning functions to different segments of the genome. For example, in a reassorted virus if one segment comes from virus A and the rest from virus B, we can see which properties resemble virus A and which virus B. Reassortment is a non-classical kind of recombination
|
||||
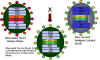 Figure 4 Reassortment of genes between an attenuated strain of influenza virus
and a new virulent strain in the formation of an attenuated influenza vaccine (link
to vaccine section) Adapted from: Treanor JJ Infect. Med. 15:714
Figure 4 Reassortment of genes between an attenuated strain of influenza virus
and a new virulent strain in the formation of an attenuated influenza vaccine (link
to vaccine section) Adapted from: Treanor JJ Infect. Med. 15:714 |
There is vaccine called Flumist (LAIV, approved June 2003) for influenza virus which involves some of the principles discussed above. The vaccine is trivalent – it contains 3 strains of influenza virus: The viruses are cold adapted strains which can grow well at 25 degrees C and so grow in the upper respiratory tract where it is cooler. The viruses are temperature-sensitive and grow poorly in the warmer lower respiratory tract. The viruses are attenuated strains and much less pathogenic than wild-type virus. This is due to multiple changes in the various genome segments. Antibodies to the influenza virus surface proteins (HA - hemagglutinin and NA - neuraminidase) are important in protection against infection. The HA and NA change from year to year. The vaccine technology uses reassortment to generate reassortant viruses which have six gene segments from the attenuated, cold-adapted virus and the HA and NA coding segments from the virus which is likely to be a problem in the up-coming influenza season. This vaccine is a live vaccine and is given intranasally as a spray and can induce mucosal and systemic immunity. A live, attenuated reassortant vaccine has recently (2006) been approved for rotaviruses (RotaTeq from Merke). Another attenuated vaccine, Rotarix (Glaxo), is in development.
|
||||
Interaction at a functional level NOT at the nucleic acid level. For example, if we take two mutants with a ts (temperature-sensitive) lesion in different genes, neither can grow at a high (non-permissive) temperature. If we infect the same cell with both mutants, each mutant can provide the missing function of the other and therefore they can replicate (nevertheless, the progeny virions will still contain ts mutant genomes and be temperature-sensitive). We can use complementation to group ts mutants, since ts mutants in the same gene will usually not be able to complement each other. This is a basic tool in genetics to determine if mutations are in the same or a different gene and to determine the minimum number genes affecting a function.
|
|||||
If double stranded DNA viruses are inactivated using ultraviolet irradiation, we often see reactivation if we infect cells with the inactivated virus at a very high multiplicity of infection (i.e. a lot of virus particles per cell) - this is because inactivated viruses cooperate in some way. Probably complementation allows viruses to grow initially, as genes inactivated in one virion may still be active in one of the others. As the number of genomes present increases due to replication, recombination can occur, resulting in new genotypes, and sometimes regenerating the wild type virus.
Defective viruses lack the full complement of genes necessary for a complete infectious cycle (many are deletion mutants) - and so they need another virus to provide the missing functions - this second virus is called a helper virus. Defective viruses must provide the necessary signals for a polymerase to replicate their genome and for their genome to be packaged but need provide no more. Some defective viruses do more for themselves.
Defective interfering particles The replication of the helper virus may be less effective than if the defective virus (particle) was not there. This is because the defective particle is competing with the helper for the functions that the helper provides. This phenomenon is known as interference, and defective particles which cause this phenomenon are known as "defective interfering" (DI) particles. Not all defective viruses interfere, but many do. Note that it is possible that defective interfering particles could modulate natural infections.
|
|||||
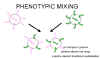 Figure 5 Phenotypic mixing between two different viruses infecting the same cell
Figure 5 Phenotypic mixing between two different viruses infecting the same cell |
If two different viruses infect a cell, progeny viruses may contain coat components derived from both parents and so they will have coat properties of both parents. This is called phenotypic mixing (figure 5). IT INVOLVES NO ALTERATION IN GENETIC MATERIAL, the progeny of such virions will be determined by which parental genome is packaged and not by the nature of the envelope. Phenotypic mixing may occur between related viruses,
e.g.
different members of the Picornavirus family, or between genetically unrelated
viruses, e.g. Rhabdo- and Paramyxo- viruses. In the latter case the two viruses
involved are usually enveloped since it seems there are fewer restraints on packaging
nucleocapsids in other viruses' envelopes
than on packaging nucleic acids in other viruses'
icosahedral capsids. |
||||
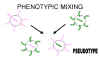 Figure 6 Phenotypic mixing to form a pseudotype
Figure 6 Phenotypic mixing to form a pseudotype |
|
||||
|
|
|||||
