| x | x | ||||||||||||
 |
|||||||||||||
| BACTERIOLOGY | IMMUNOLOGY | MYCOLOGY | PARASITOLOGY | VIROLOGY | |||||||||
|
|||||||||||||
| En Español | |||||||||||||
| SHQIP - ALBANIAN | |||||||||||||
|
Let us know what you think FEEDBACK |
|||||||||||||
| SEARCH | |||||||||||||
|
|
|||||||||||||
|
THIS CHAPTER IS IN SEVERAL PARTS USE THE NEXT>> BUTTON ABOVE TO GO TO THE NEXT PART TO CONTINUE TO VIROLOGY CHAPTER EIGHT USE LINK ABOVE |
|||||||||||||
|
LINKS TO OTHER HIV AND AIDS SECTIONS ARE AT THE BOTTOM OF THIS PAGE |
|||||||||||||
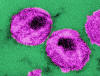 This thin-section transmission electron micrograph (TEM) depicts the
ultrastructural details of a number of ”human immunodeficiency virus” (HIV)
virus particles, or virions
This thin-section transmission electron micrograph (TEM) depicts the
ultrastructural details of a number of ”human immunodeficiency virus” (HIV)
virus particles, or virionsCDC/ Dr. A. Harrison; Dr. P. Feorino
Figure 14 - HIV structure
|
STRUCTURE OF THE VIRUS HIV is a retrovirus with a similar structure to other retroviruses (see oncogenic viruses). SURFACE STRUCTURES Viral membrane Surface glycoprotein
INTERNAL STRUCTURES Internal structural proteins Other internal proteins
For further information on retrovirus structure and replication, see oncogenic viruses Genome Since HIV has a more complex life cycle that simple retroviruses such as Rous Sarcoma Virus (RSV) and it appears that HIV can control its replication in a more complex fashion, we might expect more genetic information but this is not so. The HIV genome is 9749 nucleotides - about the same size as other retroviruses, for example RSV. The genome of HIV is more complex than RSV, however, since it has extra open reading frames that clearly code for small proteins (figure 24a). Antibodies against these small proteins are found in HIV-infected people. Some of these are protein synthesis-controlling proteins. The HIV genome has nine open reading frames (leading to nine primary translation products) but 15 proteins are made in all as a result of cleavage of three of the primary products. As we have already seen, the GAG gene and the GAG and POL genes together are
translated into large polyproteins which are then cleaved by a virus-encoded
protease that is part of the POL polyprotein.
In a addition to the nine proteins derived from GAG, POL and ENV, there are six other proteins made by HIV. Three of these are incorporated into the virus (Vif, Vpr and Nef), while the others are not found in the mature virus: Tat and Rev are regulatory proteins and Vpu indirectly assists in assembly. The genes that encode these proteins are known by three letter names that are derived as follows:
These genes encode small proteins; TAT, for example, consists of 88 amino acids. They overlap with the structural genes (especially ENV) but are in different reading frames. From the organization of the HIV genome shown in figure 24, it can be seen that some are encoded in more than one exon (unlike the structural genes) and therefore their mRNAs can be derived by alternative splicing of mRNAs for the structural proteins. Mutants in the TAT and REV genes show that both proteins are necessary for virus production.
|
||||||||||||
|
TAT TAT gene product binds to a sequence in all of the genes of HIV and positively stimulates transcription. It is thus a positive regulator of protein synthesis, including its own synthesis. |
|||||||||||||
|
REV REV binds to an element only in the mRNA for structural proteins (GAG/POL/ENV) and regulates the ratio of structural to non-structural, controlling protein (TAT/REV) synthesis. When REV levels are high, structural protein synthesis rises and the synthesis of non-structural, controlling proteins falls. Thus, REV inhibits its own production and that of TAT. The normal result is homeostasis, low or non-existent virus production and latency in the resting CD4 cell. As we have seen, there is an inherent problem in HIV's lifestyle. It uses genomic RNA as its messenger RNA. This RNA is unspliced and the nucleus has a mechanism to prevent unspliced mRNAs from leaving the nucleus and being translated. It is the function of REV to overcome this problem.
|
|||||||||||||

|
NEF NEF protein is synthesized early in infection. Despite its small size, NEF has several functions.
|
||||||||||||
|
|
VPU After activation of the T cell, the virus faces another problem: CD4 antigen and Gp120 are being made in the endoplasmic reticulum of the same cell. They are likely to bind to one another before reaching the plasma membrane and such complexes are usually targeted by the cell for degradation. To stop this unfortunate state of affairs, another of the small HIV proteins, VPU, promotes the proteolysis of the CD4 antigen of the host cell as it is made! VPU also enhances viral particle release from the host cell. How it does this is not clear but VPU forms an ion channel in the plasma membrane of the host cell and may alter the ionic composition of the cytoplasm since it conducts small ions such as Na+ and K+. It also binds to a cellular protein (VPU-binding protein or UBP) and over-expression of this protein diminishes the enhancing effect of VPU on virus release. UBP may be a negative factor for assembly that must be displaced from one of the GAG proteins before virus can assemble at the cell surface. From its ability to stimulate viral release and also to break down CD4 antigen (which are separate functions of different parts of the VPU molecule), it appears that VPU enhances the pathogenicity of the virus by increasing the number of HIV particles per cell.
VIF VIF (viral infectivity factor)
protein, which is essential for infection in vivo, may be very important
in suppressing resistance to HIV infection by the host. VIF is needed during
late stages of virus production and seems to function by suppressing innate
anti-viral activities in T cells and macrophages, the major cells that are infected in humans.
Without VIF, HIV is not infectious in primary human T cells.
VPR VPR influences the pathogenesis of HIV and is essential for infection of macrophages, and to a lesser extent of other cells. It also activates HIV LTR-promoted transcription. It causes the arrest of host cell division in the G2 stage of the cell cycle and apoptosis of the infected cell. It acts as a cytoplasmic-nucleus shuttle protein (for the pre-integration complex through the nuclear pores). VPR is found in the serum of HIV-infected patients.
|
||||||||||||
|
|
|||||||||||||
|
|||||||||||||
|
|
|||||||||||||
|
|
|||||||||||||